Custom Metadata Integration Example
This DICOM workflow demonstrates how to extract domain-specific metadata and upload it as Visual Layer custom fields, enabling powerful search and filtering capabilities. This pattern can be adapted for any specialized data type.
This example shows how custom metadata transforms basic image browsing into domain-specific dataset analysis. The same pattern applies to e-commerce, manufacturing, research, or any field with structured metadata.
Complete DICOM-to-Visual Layer Workflow
This section shows the complete end-to-end process for converting DICOM files and creating a fully searchable Visual Layer dataset with comprehensive medical imaging metadata.Extract images and metadata from DICOM files
Use the DICOM converter script to convert DICOM files to JPG images and extract comprehensive metadata to CSV format.
Create Visual Layer dataset
Upload the converted JPG images to Visual Layer to create a new dataset for analysis and exploration.
Export dataset for media_id mapping
Export your Visual Layer dataset to obtain the critical mapping between filenames and Visual Layer’s internal media_id values.
Upload DICOM metadata as custom fields
Use the DICOM metadata upload script to automatically process and upload the extracted metadata as searchable custom fields.
Step 1: Convert DICOM Files
Run the DICOM converter script to extract images and metadata. See Basic Usage section below for command examples and output details.Step 2: Create Visual Layer Dataset
Upload the converted JPG images fromconverted_images/ to Visual Layer using the web interface or API.
Step 3: Export Dataset Metadata
Export your dataset to getmetadata.json containing the filename-to-media_id mapping required for the next step.
Step 4: Upload DICOM Metadata
Use the DICOM upload script to process and upload your extracted metadata: For Cloud:Step 5: Search and Filter by DICOM Metadata
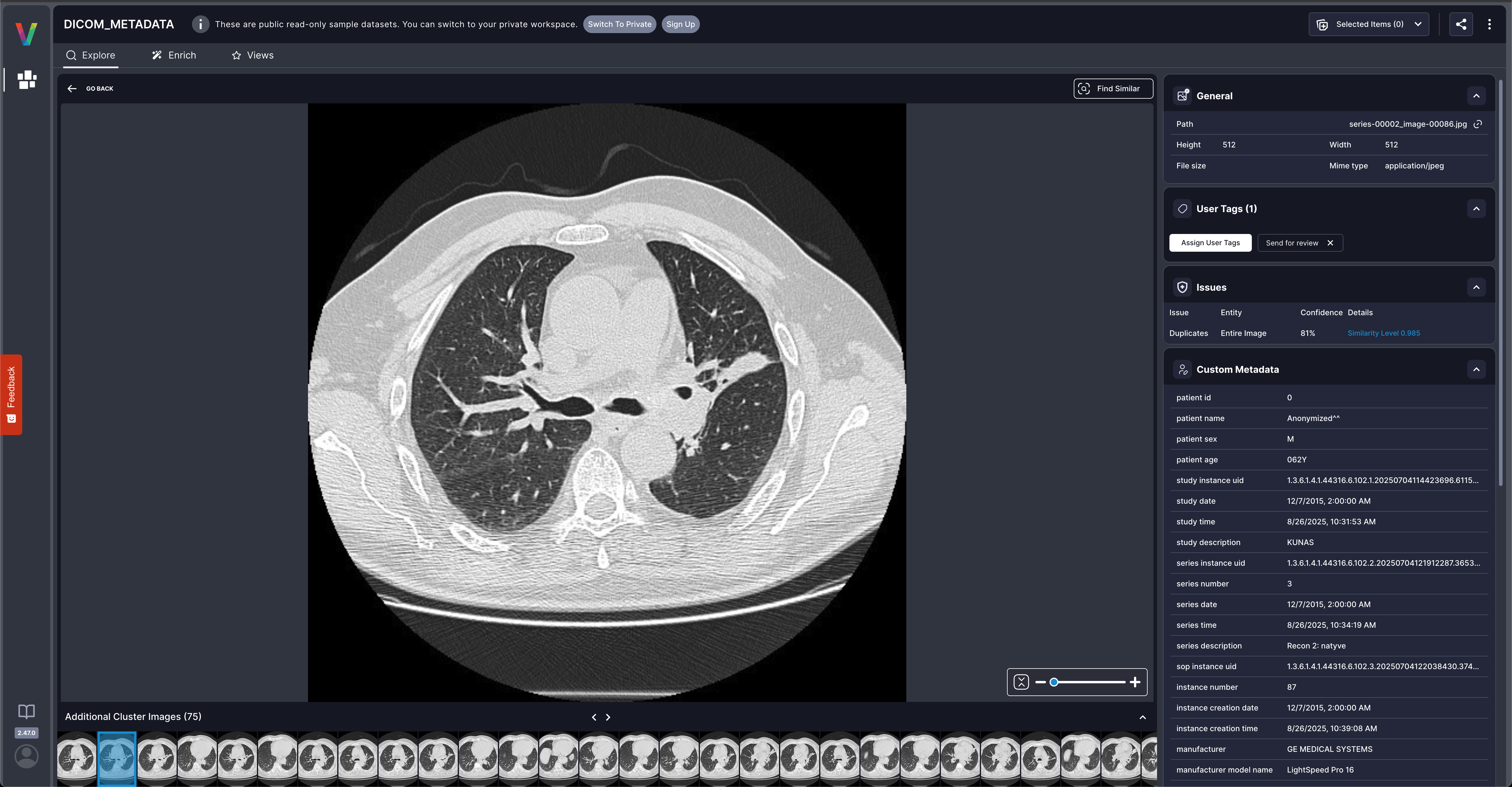
Why Custom Metadata Matters
This DICOM example shows how custom metadata transforms your Visual Layer dataset: Before custom metadata:- Browse images visually
- Basic filename-based search
- Filter by patient demographics (age, sex)
- Search by equipment manufacturer and modality
- Query by technical parameters (slice thickness, kVp)
- Group by study dates or series descriptions
- E-commerce: Product SKU, price, category metadata for inventory analysis
- Manufacturing: Quality metrics, batch numbers, inspection data for QA workflows
- Research: Experiment conditions, sample IDs, measurements for dataset curation
DICOM Converter Script
Prerequisites
- Python environment with required packages
- DICOM files organized in series directories or as individual files
- Sufficient disk space for converted images
Installation
Script Code
View Complete Script Code
Access the full DICOM converter Python script with complete implementation.
Basic Usage
converted_images/ and metadata/dicom_metadata.csv with comprehensive DICOM metadata including file info, patient data, study details, equipment parameters, and acquisition settings.
DICOM Metadata Upload Script
Visual Layer provides a customizable script for uploading DICOM metadata extracted by the converter. This script handles DICOM-specific field types and can be modified for your specific use case.Similar to the general custom metadata upload script but includes DICOM-specific field handling (date/time formats, multi-value fields).
Script Features
- Auto-detects DICOM date/time formats (YYYYMMDD, HHMMSS)
- Handles multi-value fields and complex data types
- Supports cloud and on-premises installations
- Customizable for vendor-specific DICOM tags
Script Code
View Complete Script Code
Access the full DICOM metadata upload Python script with complete implementation.
Troubleshooting
No DICOM files found:- Verify input directory contains
.dcmfiles and check file permissions
- Check verbose output for specific errors
- Verify DICOM files can be opened with standard viewers
- Ensure dataset ID is correct and dataset is in READY state
- Use
--resumeflag to continue interrupted uploads
Adapting This Pattern
The DICOM scripts can be customized for your specific use case:- Modify field extraction to capture vendor-specific tags
- Add preprocessing for specialized protocols
- Extend to other formats by adapting the extraction logic
Related Resources
- Custom Metadata API Guide - Manual API workflow
- Custom Metadata Upload Script - General pattern for other data types
- Creating Datasets - Dataset creation guide